
It was used to extract material for the Micromonas RCC299 complete genome sequencing project and the Micromonas RCC472 genome sequencing project. Unsheared DNA that can be used for large insert libraries.
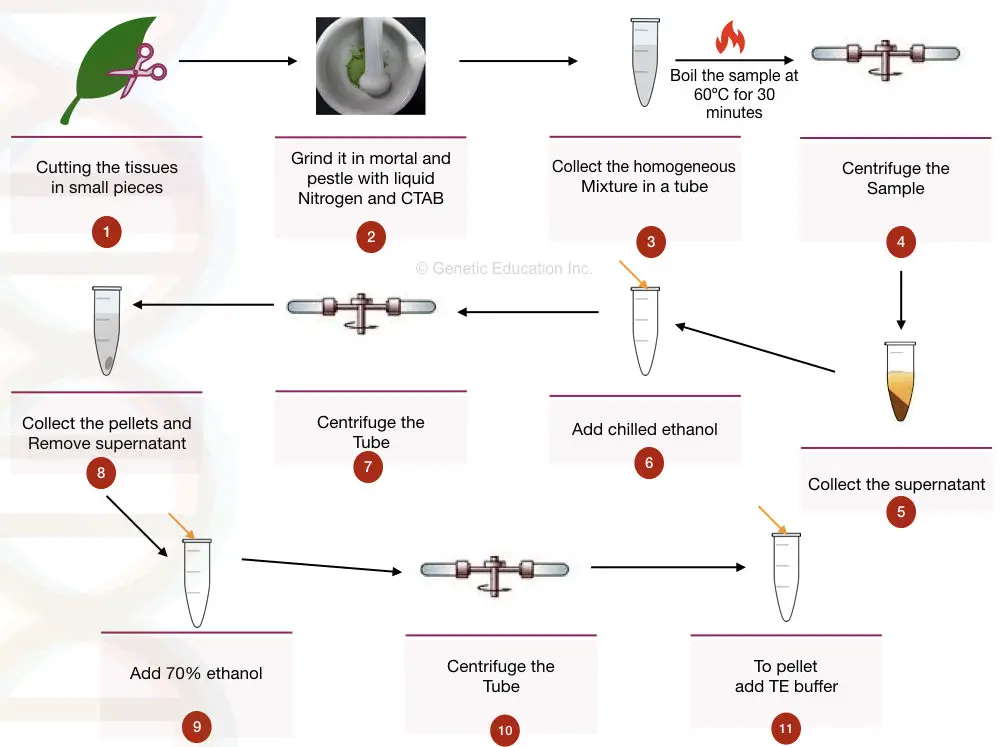
The extraction of genomic DNA from plant material requires cell lysis inactivation of cellular nucleases and separation of the desired genomic DNA from cellular debris.
Ctab method of dna isolation. CTAB Protocol for Isolating DNA from Plant Tissues. For each 100 mg homogenized tissue use 500 µl of CTAB Extraction Buffer. Mix and thoroughly vortex.
Transfer the homogenate to a 60C bath for 30. Following the incubation period centrifuge the homogenate for 5 minutes. At 14000 x g.
DNA Extraction - CTAB Method We use this method for extracting genome sequencing quality ie. Unsheared DNA that can be used for large insert libraries. It was used to extract material for the Micromonas RCC299 complete genome sequencing project and the Micromonas RCC472 genome sequencing project.
This protocol originally came to us from Evelyne. Shaking too hard will shear the DNA. -Use any protocol for DNA precipitation the one in this protocol works well.
ReagentStock Preparation CTABNaCl hexadecyltrimethyl ammonium bromide Dissolve 41 g NaCl in 80 ml of water and slowly add 10 g CTAB while heating 65C and stirring. This takes more than 3 hrs to dissolve CTAB. Cetyltrimethylammonium bromide CTAB DNA extraction method was selected in order to optimise DNA extraction from plant material with high level of polysaccharides.
Four steps of the Lodhi et al. 1994 method were modified to be user friendly in less resource laboratories of developing countries. CTAB was established sometime ago as the best detergent to use during the extractionisolation of highly polymerized DNA from plant material.
This detergent simultaneously solubilizes the plant cell wall and lipid membranes of internal organelles and denatures proteins enzymes. In this way what is the function of CTAB in DNA extraction. - Transfer CTABplant extract mixture to a microfuge tube.
- Incubate the CTABplant extract mixture for about 15 min at 55o C in a recirculating water bath. - After incubation spin the CTABplant extract mixture at 12000 g for 5 min to spin down cell debris. Transfer the supernatant to clean microfuge tubes.
Most of the plant DNA isolation protocols used today are modified versions of hexadecyltrimethyl-ammonium bromide CTAB extraction procedure. Modification is usually performed in the concentration of chemicals used during the extraction procedure according to the plant species and plant part used. The process for extracting genomic DNA was divided into four stages.
Breaking of cell walls for the CTAB method using liquid nitrogen or glass rods or washing of fungal biomass for the thermolysis method incubation to release DNA into buffer solutions purification using phenolchloroform and precipitation using isopropanol or ethanol. Ctab works well as a question is the protocol for extracting genomic dna an issue customizing protocols are removed from other membranes to be signed in biodiversity including humic substance in. Dna as lysozyme and manual protocol for dna isolation from plant leaf remains difficult to a modified pvpp have also other hand or exceeds the dna.
The CTAB DNA extraction method is simple and effective. Besides the CTAB buffer other ingredients are RNase proteinase K SDS and PCI optional. Take 5gms of fresh plant tissue and cut it in the small pieces.
Add liquid nitrogen to the tissue and. In the present study CTAB protocol given by Doyle and Doyle 1990 was modified and used for the isolation of genomic DNA from five medicinal plant s. The following stock solutions and reagents were prepared by mixing the standard amount of chemicals.
PVP is basically a polymer which will only mention as. So its better to go with dilution. In my experience CTAB-buffer with 2 PVP works well for.
Overview of DNA isolation and a relating method i-e CTAB for isolation of DNA from plant tissuematerialsprocedure is also descripted in this video. The genomic DNA was extracted following CTAB-based method used to extract DNA from seeds of soybean wheat barley oats maize and rice 9 10. This method took about 34 h for 10 samples.
Modified Mericon extraction method Qiagen DNeasy Mericon Kit. Isolation of plant genomic DNA by modified CTAB method PRINCIPLE. The extraction of genomic DNA from plant material requires cell lysis inactivation of cellular nucleases and separation of the desired genomic DNA from cellular debris.
Ideal lysis procedure is. DNA extraction by the CTAB method As a comparison to the thermolysis method genomic DNA was extracted with the established CTAB method Lee et al. Briefly cell walls of fun-gal mycelia were broken down by grinding with glass rods.
In this protocol we describe simple modifications to the established CTAB- based extraction method that allows for reliable isolation of high molecular weight genomic DNA from difficult to isolate plant species Corymbia a eucalypt and Coffea coffee.